Linear regression
Here, . Therefore, we can re-write as:
In Bayesian linear regression, we assume as data and as parameters. Therefore, the likelihood is as follows:
Using Bayes rule, the posterior distribution can be written as:
Now, we can assume and to be independent of each other. We can further assume, all the elements of are independent. Therefore, prior can be written as:
Prior Distribution
When , then the MAP estimate is ridge regression.
When , i.e. Laplacian distribution, then the MAP estimate is the lasso regression.
Linear Regression using sklearn
First we import all the necessary functions:
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
sns.set_style("whitegrid")
sns.set_context("poster")
from sklearn.linear_model import LinearRegression
from sklearn.linear_model import Lasso, LassoCV
from sklearn.linear_model import Ridge, RidgeCV
import pymc3 as pm
We are generating some data points for the function and performing regular linear regression, Ridge regression and Lasso regression.
Ridge Regression
Lasso Regression
In our simulation, for Ridge and Lasso regression, we are intentionally setting some extreme values of so that we can see the feature selection characteristics of these regression. See this blog post for a very nice explanation of the behavior of Ridge and Lasso regression.
Note that, sklearn Ridge and Lasso doesn’t have lambda parameter, instead it has alpha parameter. My guess is, for Ridge lambda and alpha are the same. However, for Lasso, the description here is a bit confusing. After some trial and error, my suspicion is:
alpha = number_of_samples x lambda
This is important for this analysis, since in the next section, we need to choose proper prior for our MCMC simulation depending on this value.
size = 1000 # number of data poins
np.random.seed(seed=1)
X_seed = np.random.normal(0, 1, size)
X1 = X_seed + np.random.normal(0, .1, size)
X2 = X_seed + np.random.normal(0, .1, size)
X3 = X_seed + np.random.normal(0, .1, size)
sigma = 1
noise = np.random.normal(0, sigma, size)
Y = 1 * X1 + 1 * X2 + 1 * X3 + noise
X = np.array([X1, X2, X3]).T
# Linear Regression
lr = LinearRegression()
lr.fit(X,Y)
print 'Linear model : {}'.format(lr.coef_)
# Ridge Regression (lamba = 1000)
ridge = Ridge(alpha=1000)
ridge.fit(X,Y)
print 'Ridge model : {}'.format(ridge.coef_)
# Lasso Regression
# From the description here http://scikit-learn.org/stable/modules/generated/sklearn.linear_model.Lasso.html
# I am a little confused
# my guess is lambda = alpha * n_samples
lasso = Lasso(alpha=1.)
lasso.fit(X, Y)
print 'Lasso model : {}'.format(lasso.coef_)
Linear model : [ 1.16266223 0.98860861 0.8087923 ]
Ridge model : [ 0.73644275 0.72897954 0.73591734]
Lasso model : [ 0.89910385 0. 1.02094343]
MCMC Bayesian Regression
First, we assumed uniform prior for the regression co-efficients and perform the simulation:
# using uniform prior to find the co-efficient
w_min = -10
w_max = 10
with pm.Model() as uni:
w1 = pm.Uniform('w1', lower=w_min, upper=w_max)
w2 = pm.Uniform('w2', lower=w_min, upper=w_max)
w3 = pm.Uniform('w3', lower=w_min, upper=w_max)
wTx = w1*X1 + w2*X2 + w3*X3
y = pm.Normal('y', mu=wTx, sd=sigma, observed=Y)
stepper=pm.Metropolis()
traceuni = pm.sample(100000, step=stepper)
69%|██████▉ | 69407/100500 [00:48<00:21, 1434.44it/s]
Once finished, we can plot and get summary statistics as follows:
pm.traceplot(traceuni[20000::50])
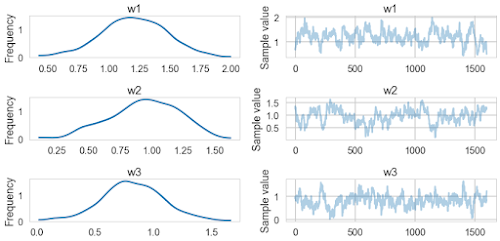
pm.summary(traceuni[20000::50])
w1:
Mean SD MC Error 95% HPD interval
-------------------------------------------------------------------
1.207 0.268 0.023 [0.609, 1.688]
Posterior quantiles:
2.5 25 50 75 97.5
|--------------|==============|==============|--------------|
0.644 1.033 1.208 1.397 1.733
w2:
Mean SD MC Error 95% HPD interval
-------------------------------------------------------------------
0.938 0.273 0.025 [0.399, 1.407]
Posterior quantiles:
2.5 25 50 75 97.5
|--------------|==============|==============|--------------|
0.400 0.764 0.952 1.147 1.408
w3:
Mean SD MC Error 95% HPD interval
-------------------------------------------------------------------
0.819 0.262 0.022 [0.304, 1.371]
Posterior quantiles:
2.5 25 50 75 97.5
|--------------|==============|==============|--------------|
0.275 0.650 0.812 0.991 1.355
We can also try to calculate the MAP estimate of the co-efficients. This should be the same as ML estimate in the last section.
with uni:
uni_map = pm.find_MAP()
print(uni_map)
logp = -1,447.3, ||grad|| = 4.4401: 100%|██████████| 16/16 [00:00<00:00, 1157.33it/s]
{'w3_interval__': array(0.16224552703292178), 'w1_interval__': array(0.23723549815233705), 'w3': array(0.8094527703753798), 'w2': array(0.9728674588105903), 'w1': array(1.1806453839461994), 'w2_interval__': array(0.19519086212979697)}
Bayesian Prior - Ridge Regression
Here, we are making sure that the prior distribution of the co-efficients have zero mean and variance so that it gives us the same MAP estimate as in sklearn Ridge regression earlier.
alpha = 1000. # we used this value for sklearn Ridge estimation
lambda_1 = alpha
sigma = 1
sd = np.sqrt((sigma**2)/lambda_1)
print(sd)
with pm.Model() as ridge:
w1 = pm.Normal('w1', mu=0, sd=sd)
w2 = pm.Normal('w2', mu=0, sd=sd)
w3 = pm.Normal('w3', mu=0, sd=sd)
wTx = w1*X1 + w2*X2 + w3*X3
ys = pm.Normal('ys', mu=wTx, sd=sigma, observed=Y)
stepper=pm.Metropolis()
traceridge = pm.sample(100000, step=stepper)
0.0316227766017
100%|██████████| 100500/100500 [01:11<00:00, 1402.93it/s]
pm.traceplot(traceridge[20000::50])
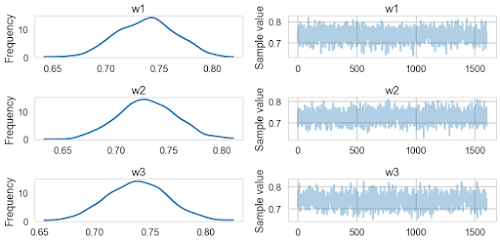
pm.summary(traceridge[20000::50])
w1:
Mean SD MC Error 95% HPD interval
-------------------------------------------------------------------
0.738 0.028 0.001 [0.683, 0.789]
Posterior quantiles:
2.5 25 50 75 97.5
|--------------|==============|==============|--------------|
0.684 0.718 0.739 0.757 0.791
w2:
Mean SD MC Error 95% HPD interval
-------------------------------------------------------------------
0.729 0.027 0.001 [0.675, 0.779]
Posterior quantiles:
2.5 25 50 75 97.5
|--------------|==============|==============|--------------|
0.676 0.711 0.729 0.748 0.783
w3:
Mean SD MC Error 95% HPD interval
-------------------------------------------------------------------
0.738 0.027 0.001 [0.687, 0.794]
Posterior quantiles:
2.5 25 50 75 97.5
|--------------|==============|==============|--------------|
0.684 0.719 0.738 0.756 0.792
We can also estimate the MAP values for the co-efficients.
with ridge:
mapridge = pm.find_MAP()
mapridge
{'w1': array(0.7377441385493746),
'w2': array(0.7297896623054996),
'w3': array(0.7369597481542057)}
Note that, these values are similar to the values obtained from sklearn Ridge.
Laplacian Prior - Lasso Regression
Note that, I am guessing the value of here due to the mismatch between the Lasso equation and sklearn implementation. The conversion in the first few line ensure that we select proper for our Laplacian priors.
alpha=1. # we used this value for sklearn Lasso
lambda_1 = alpha * size # my guess here is, lambda = alpha x num_samples
sigma = 1.
b = sigma**2 / lambda_1
print(b)
with pm.Model() as lasso:
w1 = pm.Laplace('w1', mu=0, b=b)
w2 = pm.Laplace('w2', mu=0, b=b)
w3 = pm.Laplace('w3', mu=0, b=b)
wTx = w1*X1 + w2*X2 + w3*X3
ys = pm.Normal('ys', mu=wTx, sd=sigma, observed=Y)
stepper=pm.Metropolis()
tracelaplace = pm.sample(100000, step=stepper)
0.001
100%|██████████| 100500/100500 [01:14<00:00, 1343.27it/s]
pm.traceplot(tracelaplace[20000::50])

pm.summary(tracelaplace[20000::50])
w1:
Mean SD MC Error 95% HPD interval
-------------------------------------------------------------------
0.859 0.241 0.019 [0.412, 1.322]
Posterior quantiles:
2.5 25 50 75 97.5
|--------------|==============|==============|--------------|
0.389 0.695 0.863 1.034 1.304
w2:
Mean SD MC Error 95% HPD interval
-------------------------------------------------------------------
0.179 0.141 0.009 [-0.001, 0.453]
Posterior quantiles:
2.5 25 50 75 97.5
|--------------|==============|==============|--------------|
0.007 0.068 0.145 0.255 0.551
w3:
Mean SD MC Error 95% HPD interval
-------------------------------------------------------------------
0.890 0.237 0.020 [0.461, 1.371]
Posterior quantiles:
2.5 25 50 75 97.5
|--------------|==============|==============|--------------|
0.421 0.725 0.891 1.053 1.345
We can also find the MAP estimate:
with lasso:
maplasso = pm.find_MAP()
maplasso
{'w1': array(0.9500293027663775),
'w2': array(1.3861762347993195e-09),
'w3': array(0.9796505403007957)}
Note that, this values are similar to the values estimated using sklearn Lasso earlier.















